Progress Update: Sep-Nov 2025
Regulome profiling, new paths to the androgen receptor, and oncogenic fusion proteins
Progress continues at Talus Bio as we head into the holiday season. We’re close to having our first clinic-bound molecule for an “undruggable” regulome target in hand and we’ll share an update on that next year.
Today instead, I want to highlight a couple of newer programs that showcase the power of combining regulome profiling with emerging AI models for drug discovery.
Most drug discovery approaches are reductionist by necessity.
If you’re screening millions or billions of compounds to find a hit, then you need to keep the assay simple. Usually that means measuring one thing at a time (does a compound bind its target? degrade a protein?). That also means engineering an artificial system, destroying the native biological state of the drug target. This narrow view works fine for simple mechanisms, the “druggables”. But most of these low-hanging fruits have been picked clean off the tree.
At Talus, we built a technology that profiles the entire native regulome.
Native means that each time that we test a compound, we see how it affects everything that controls the genome in an unmodified human cell. That includes transcription factors, chromatin regulators, remodelers, the RNA processing machinery, and the complex interplay between them all. Most technologies in biology have a Schrodinger’s Cat problem where measuring the system, by definition, requires disrupting it. What we get here is a unique opportunity to take high-resolution snapshots of what the regulome is doing without disturbing it, and see how it changes over time.
These snapshots open up a new kind of therapeutic opportunity.
Instead of screening compounds blindly in the lab with simple assays, we now have enough data to train AI to virtually screen billions of compounds, then bring the strongest hits into the lab to test, validate, and improve our models. Virtual hits are validated directly in native biological systems. This happens on the order of weeks.
The most important “undrugged” targets in complex disease are within the regulome and work through unpredictable, deeply interconnected mechanisms. Now, we can finally start to untangle how this system works, and I’ll share two recent examples.
A Protein-RNA Molecular Glue for Castration-Resistant Prostate Cancer
Six months ago, we initiated a second program with our regulome profiling engine targeting the androgen receptor (AR) in prostate cancer.
AR drives prostate cancer. The entire treatment paradigm is built around blocking it. First with hormone therapy, then with more potent AR inhibitors like enzalutamide and apalutamide. But prostate cancer is determined to reactivate AR any way it can. We see splice variants and mutations that block drug binding, amplification of the AR gene itself, even tumors that synthesize their own testosterone to boost AR signaling. The cancer works extraordinarily hard to keep AR signaling alive, and eventually it succeeds in most patients.
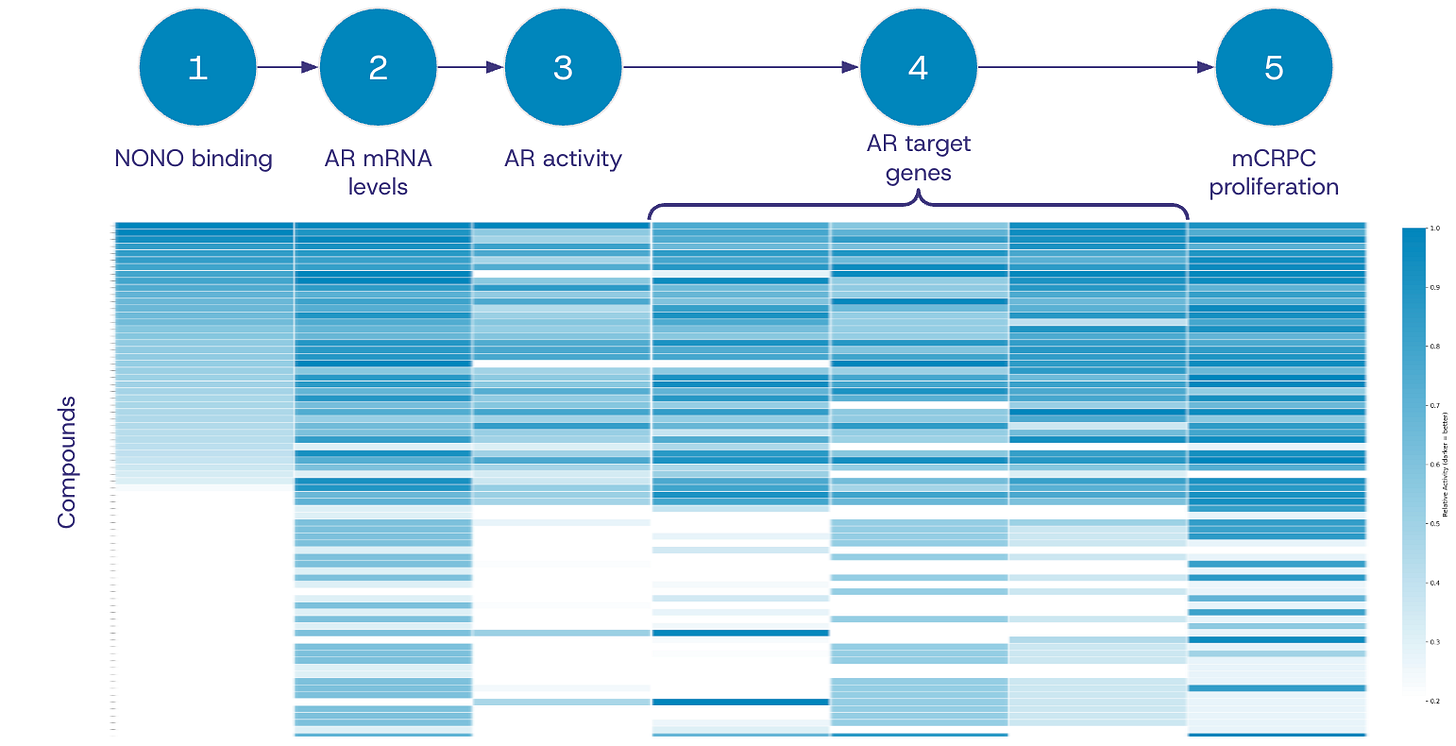
Our platform identified a way to cut off AR signaling before the protein is even made with a molecule that sticks to a regulome protein (NONO) and prevents the AR mRNA from maturing and ever being translated into AR protein, depleting AR protein from binding to the genome.
We started this program nine months ago, and regulome profiling has proven essential. Unlike a traditional screen where you’re measuring one binding event or one cellular readout, we can directly visualize:
Compound binding directly to the NONO protein
How that impacts levels of AR mRNA
AR levels in the cell and directly on DNA, including all mutants or splice variants
Which off-target proteins are being bound and altered
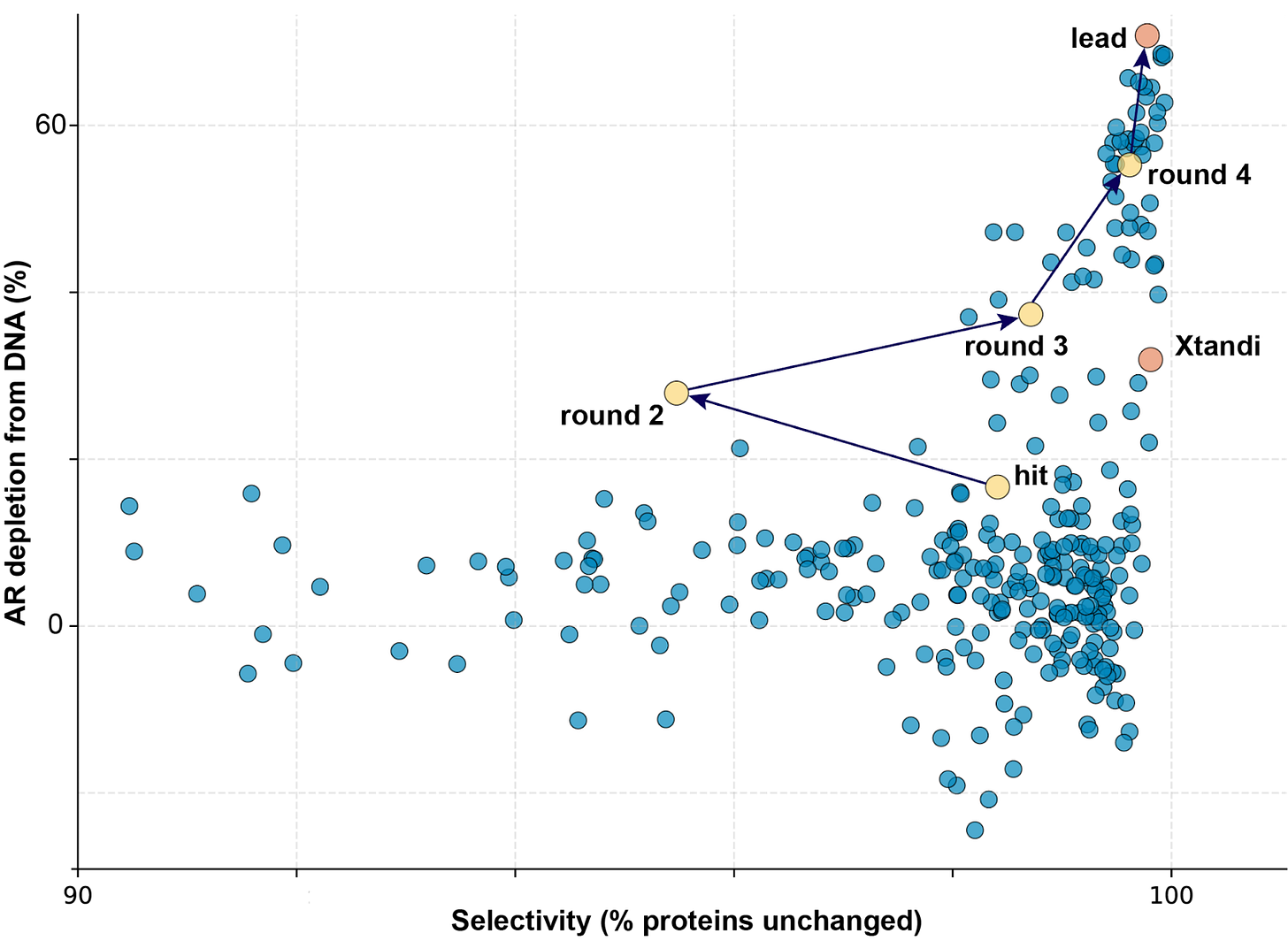
This technology lets us iterate incredibly quickly. Starting from our original hit compound, we’ve now tested over 350 versions and nominated TAL20231 as a lead compound that is currently in in vivo evaluation. Compared to our original hit, this compound has:
5-fold improvement in potency
10-fold enhanced selectivity with very few off-target binding events
95% target engagement at the NONO/AR mRNA complex
A 10-fold improvement in metabolic stability
Longer microsomal stability in both human (>145 min vs 26 min) and mouse (>49 min vs 16 min)
Room to grow as we progress through lead optimization (MW of 416 g/mol)
The speed here is remarkable.
Six months from program start to a compound with this profile was only possible because regulome profiling gave us direct-to-biology mechanistic readouts at each optimization cycle.
Unlocking a Fusion Transcription Factor with AI regulome models
Beyond optimizing our programs in flight, our platform is unlocking new starting points across historically “undruggable” target space.
These targets can’t be solved with AlphaFold or structure-based design because many regulome proteins like transcription factors work through dynamic, context-dependent interactions across, not static binding pockets.
So we trained our own AI model.
This is possible now, with our regulome data atlas hitting an inflection with over 100M data points. The result is a multi-modal AI model combining our unique regulome data with state-of-the-art protein and chemical foundation models to run virtual screens for “undruggable” targets.
Already, our platform has delivered chemical starting points for 68% of all previously “undruggable” transcription factor families.
One example I’m particularly excited about is an oncogenic fusion transcription factor that drives a type of aggressive childhood tumor. It’s a perfect example of an “undruggable” target where two transcription factors are fused into one new mutant protein that activates genes that drive uncontrolled growth. These fusions have no enzymatic activity to inhibit, no clear binding pocket for a traditional small molecule, and they work by re-wiring the cell’s transcriptional machinery in complex ways.
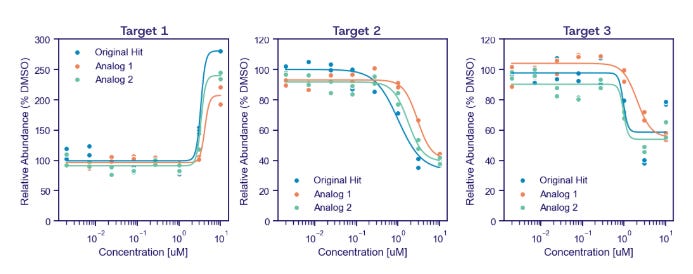
Through lab-in-the-loop active learning, we converged on a series of molecules that directly disrupted the fusion and shut off its downstream gene targets. Here’s an example of 3 members in the series modulating 3 downstream gene targets in the expected directions.
We’re still early here, focused on hit expansion and establishing SAR. But it demonstrates the platform can generate chemical starting points even for targets most drug hunters have thrown in the towel for.
What’s Next
We’re energized every day by peering into biology other platforms can’t see, and translating directly into unlocking and accelerating new therapeutic opportunities.
Join our tech access program, or join our team.
Or see what else we’ve been up to


The leap from hit identification to lead optimization in just 6 months for TAL20231 is really solid work. What stands out is how the native regulome profiling sidesteps the Schrodinger's Cat issue most biochem assays face. Directly visualizing compound binding to NONO while tracking AR mRNA levels and genomic occupancy in parallel is kinda the whole point of systems-level drug discovery. The 68% hit rate on previously undruggable TF families sounds ambitous but the fusion TF data suggests the model is actually learning somethig beyond brute force pattern matching.